Molecular Profiles Overview¶
CIViC Molecular Profiles are combinations of one or more CIViC variants. Most Molecular Profiles are “Simple” Molecular Profiles comprised of a single variant. In most cases, these can be considered equivalent to the CIViC concept of a Variant. However, increasingly clinical significance must be considered in the context of multiple variants simultaneously. Complex Molecular Profiles in CIViC allow for curation of such variant combinations. Regardless of the nature of the Molecular Profile (Simple or Complex), it must have a Predictive, Prognostic, Predisposing, Diagnostic, Oncogenic, or Functional clinical relevance to be entered in CIViC.
The co-occurrence and mutual exclusivity of mutations in cancer are gaining clinical relevance. Double Hit Lymphoma (DHL) is characterized by combinations of mutations in MYC, BCL2, or BCL6. Trastuzumab resistance in HER2 overexpressing breast cancer may be induced by mutations in PIK3CA. Resistance to Imatinib may be conferred by mutations to BCR-ABL fusions (e.g. BCR::ABL + ABL F317L). Recent studies have indicated that checkpoint inhibitor therapy targeting PDL1 may be more effective in the absence of strong drivers like ALK Fusion or EGFR Mutation.
Past versions of the CIViC data model had Gene as a top level entity, associated to one or more Variants, with literature curated Evidence Items (EIDs) supporting each variant (Figure 1). Evidence Items could only be curated for variants associated with a single gene, so curation of evidence for an entity like DHL would not be possible. Molecular Profiles (MPs) were introduced to address this shortcoming of the data model.
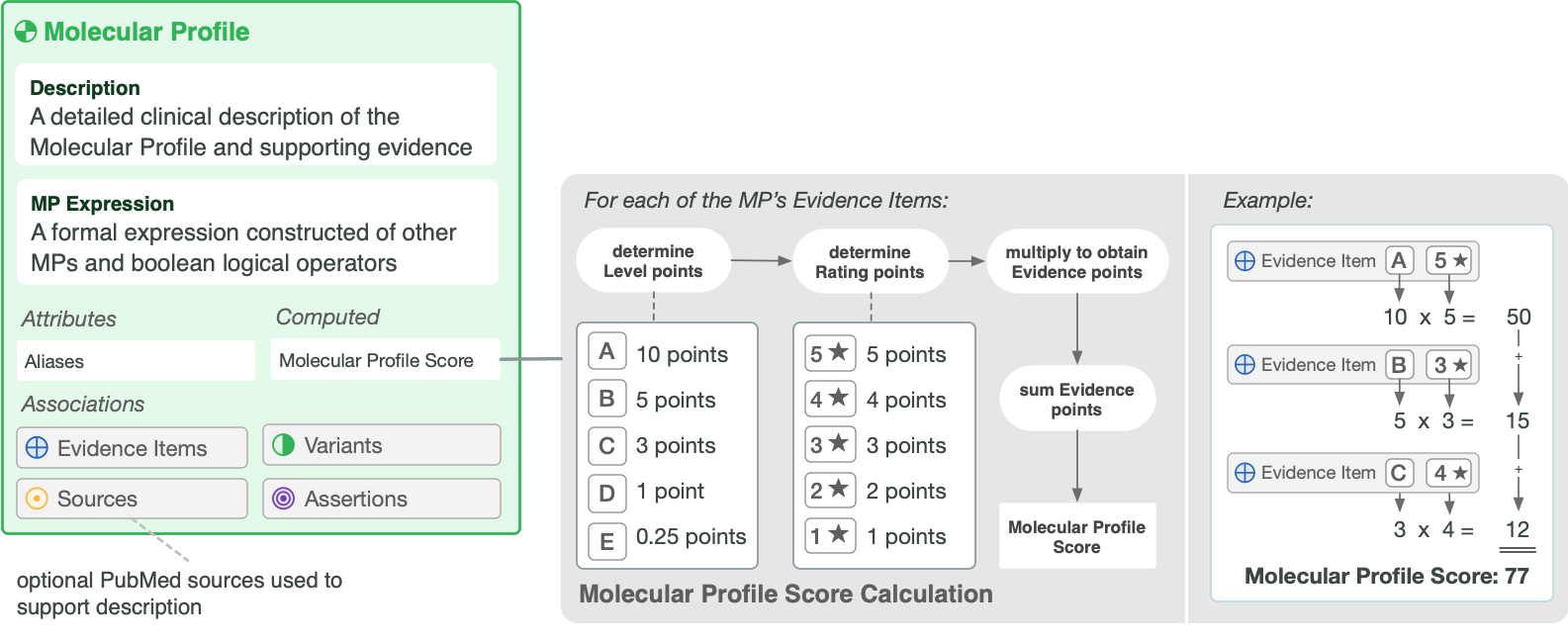
Figure 2 shows the attributes of a Molecular Profile, its associations with other CIViC entities, and how its Score is computed.
Consider the following case study:
A 42 year old man with non-smoking history presented with EGFR L858R positive lung adenocarcinoma. He was given various treatments including erlotinib and gefitinib and progressed four years later. Biopsy revealed EGFR T790M mutation, and osimertinib treatment was given. Partial response was seen, and progression occurred after 13 months. BRAF V600E mutation was found in progressed tumor cells, but was not reported before osimertinib resistance.
This case describes a lung cancer patient with EGFR L858R, EGFR T790M, and BRAF V600E mutations gaining resistance to osimertinib. We define the following Molecular Profile (MP) to describe the relevant patient genotype of driver and resistance variants described in the case study:
EGFR L858R AND EGFR T790M AND BRAF V600E
Earlier versions of CIViC did not allow for evidence to be associated with collections of variants across different genes such as the MP above. To address this need, the CIViC data model has been updated to include MPs which allow the association of clinical evidence to complex combinations of multiple variants across different genes. The MP consists of a combination of one or more variants. Variants are placed in combinations connected by “AND”, “OR”, “AND NOT”, or “OR NOT”. These relationships may be further defined by parenthesis, as in DHL which has variants in MYC and either BCL2 or BCL6:
MYC Rearrangement AND (BCL2 Rearrangement OR BCL6 Rearrangement)
The specific absence of a variant in the MP is supported by AND NOT. An example of this is a Molecular Profile which may be indicated for checkpoint inhibitor therapy:
PDL1 Expression AND NOT (EGFR Mutation OR ALK Fusion)
Which is equivalent to:
PDL1 Expression AND NOT EGFR Mutation AND NOT ALK Fusion
Currently no specific form among equivalent MPs is enforced. Alternate forms for an MP should appear as aliases on the MP page. Curators should strive for the shortest and most intuitive representation of the MP. Evidence Items (EIDs) in the new model are associated with MPs instead of directly with variants. There are two types of MP, the simple MP which contains only one variant, and the complex MP which contains two or more variants (Figure 3). The variants in a complex MP can be associated with one or more genes.

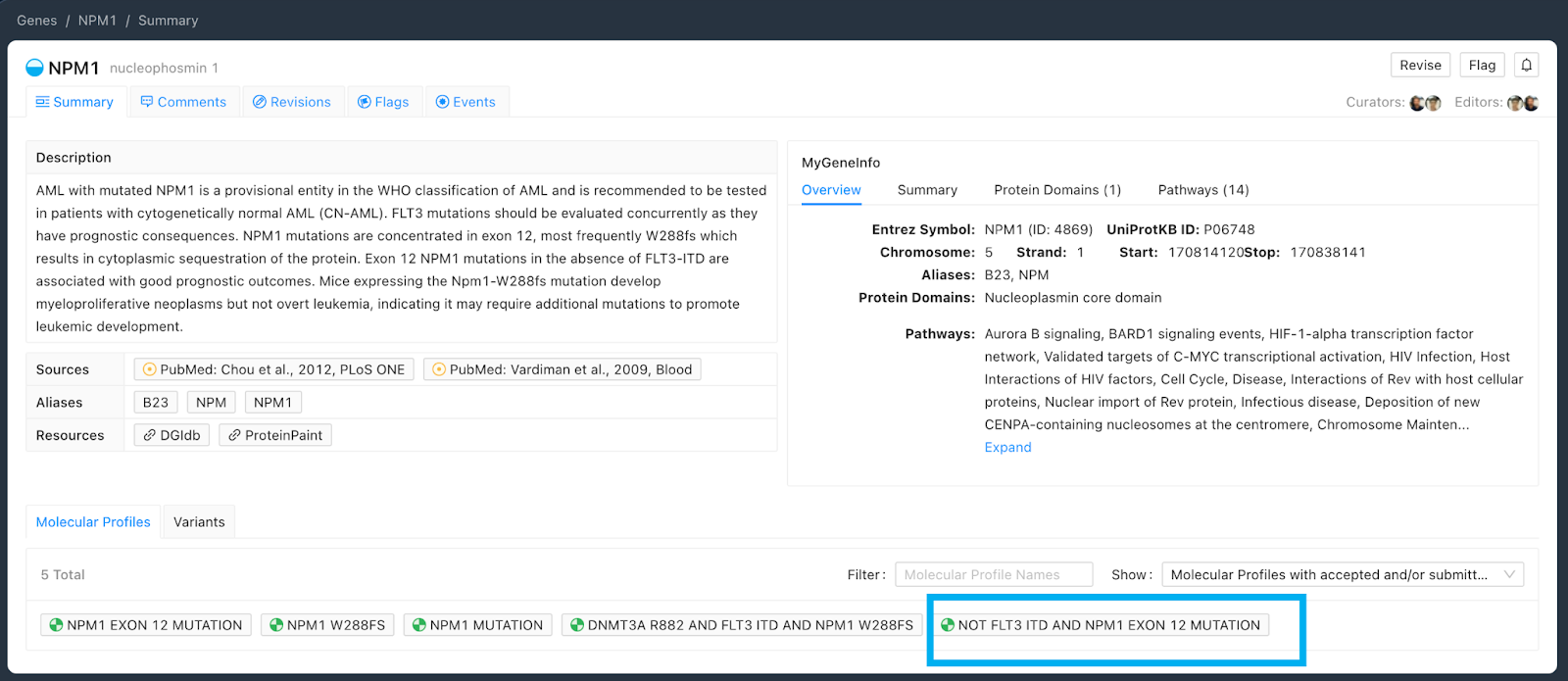
A complex MP which has two or more variants can also be thought of as containing smaller MPs within it. Therefore, in the example Molecular Profile “NPM1 Exon 12 Mutation AND NOT FLT-3 ITD” (Figure 4), we see that Evidence Items can be associated with the complex MP as well as to the simple MPs of which the complex MP is comprised of:

Molecular Profiles associated with a gene are visible on the gene page (Figure 5). The list contains the simple MPs which are associated only to the given gene, as well as complex MPs which can be composed of multiple variants, including variants associated with other genes as well as the given gene. Note that the Molecular Profile “NPM1 Exon 12 Mutation AND NOT FLT3 ITD” is displayed in the order with FLT3 ITD first in Figure 5.
Molecular profile naming follows a structure where the gene is named first, followed by the specific variant name, and for complex MPs this pattern is followed as well, but linked with AND, OR, and NOT. Therefore the gene EGFR and the variant L858R will together comprise the Molecular Profile EGFR L858R.
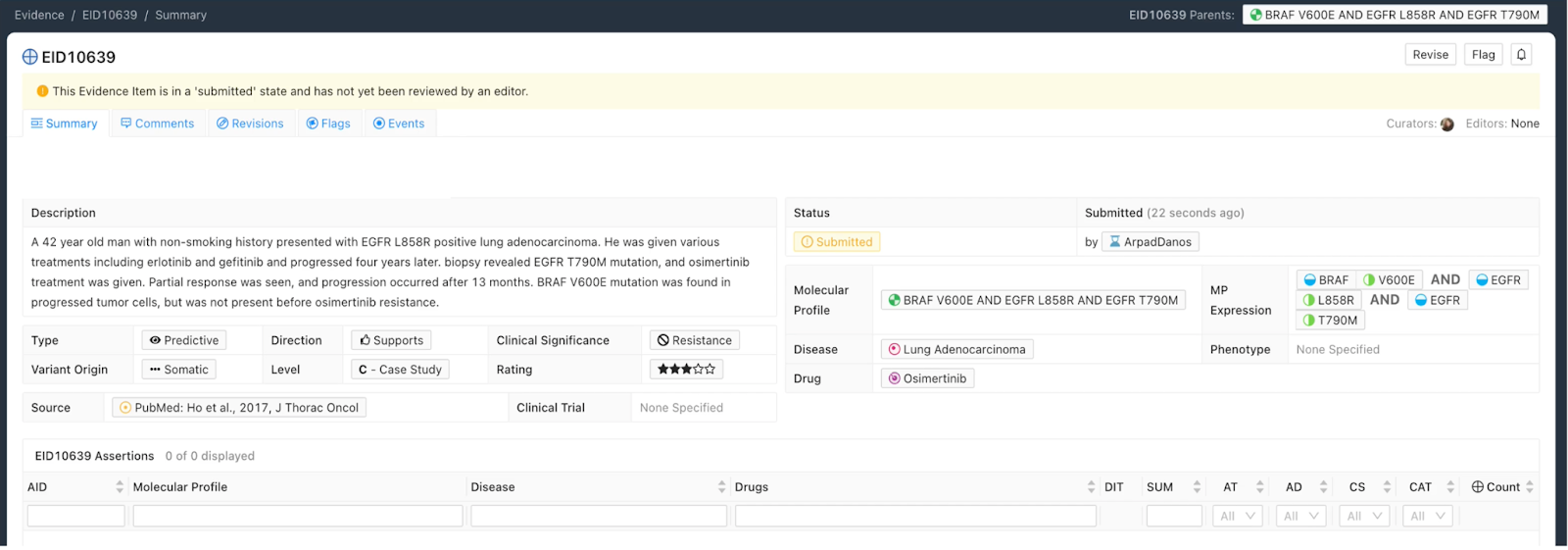
Evidence Items based on complex Molecular Profiles are drawn from the same six evidence types and contain the same structured fields to be filled out by the curator as Evidence Items based on simple MPs, or Evidence Items based on single gene variants from older versions of CIViC. The case study mentioned above has been curated into an evidence item and is seen in the example EID in Figure 6.
Molecular Profile Attributes
Attribute |
Description |
Source |
|---|---|---|
Name |
Name of the Molecular Profile. This is assembled automatically from the component gene/variant parts. |
CIViC |
Description |
User-defined description of the clinical relevance of this molecular profile. |
CIViC |
Alias |
Alternative names for this Molecular Profile |
CIViC |
Sources |
A list of PubMed IDs referring to evidence supporting statements made in the Molecular Profile description. Source descriptions (e.g. ‘Weisberg et al., 2007, Nat. Rev. Cancer’) are pulled from the PubMed database at the time of submission, and are not editable. |
CIViC (PubMed) |
Molecular Profile Score |
The Molecular Profile Score assesses the quality and quantity of evidence submitted for each molecular profile. The Molecular Profile Score is calculated by adding all Evidence Item Scores for each variant. The Evidence Item Score is calculated by multiplying the evidence level (A=10 points, B=5 points, C=3 points, D=1 point, E=0.25 points) by the evidence rating (Each Star = 1 point). |
CIViC |